A method to identify cell-free biomarkers of cancers and other diseases
A cost-effective method to discover biomarkers for a variety of cancers that can be measured in a blood sample and can inform risk, diagnosis, prognosis and predict responses to therapy.
Applications
- Biomarker discovery for cancers and other diseases
- Identification of conserved inter-species disease biomarkers
Key Benefits & Differentiators
- Robust biomarker discovery: Method identifies signatures containing multiple biomarkers and accounts for the heterogeneity between cancer cells in the same tumor and between patients with the same cancer.
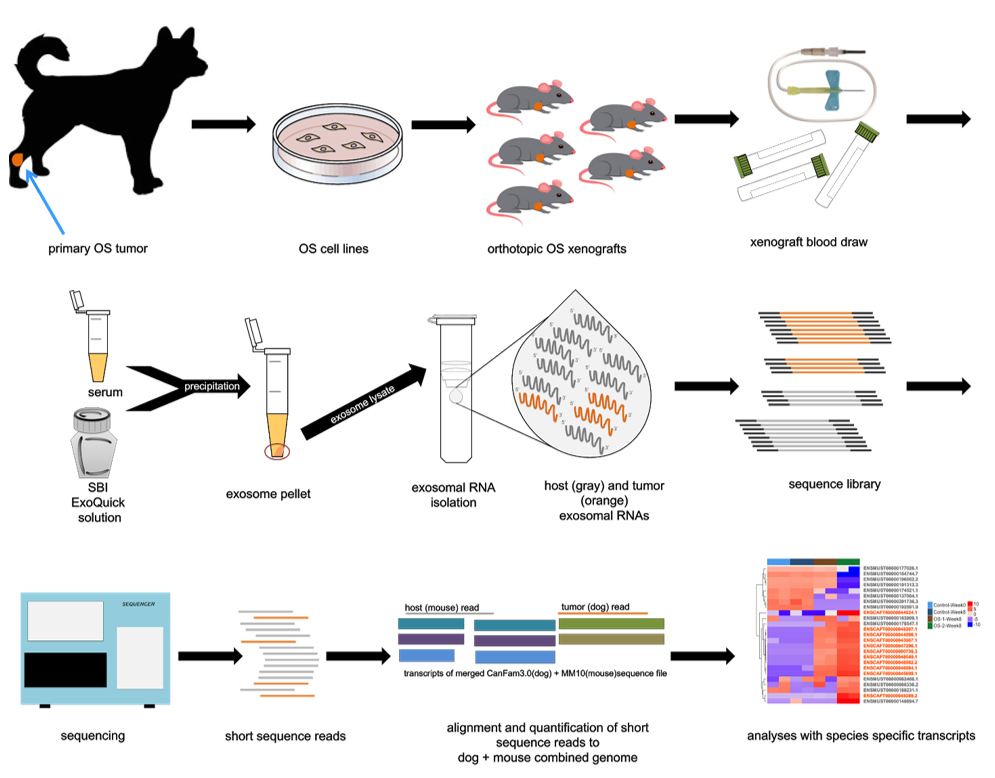
- Host vs. disease biomarker identification: Approach uses xenograft models, allowing discrimination between biomarker changes in the host and in the disease.
- Easy and cost-effective sample collection: Method identifies RNA biomarkers in exosomes wich can be recovered from a blood sample.
- Applicable for diagnostic or prognostic biomarker identification
The value of precise and accurate biomarkers
Precision diagnostics are among the unmet medical needs that are required to achieve efficient and cost-effective personalized health care. Specifically, robust methods are needed to uncover biomarkers that serve as accurate predictors of risk, early detection, prognosis, and companion diagnostics for individualized therapies. Currently, biomarker discovery results in identification of single markers. While initially promising, these standalone markers often fail due to the heterogeneity observed within tumors in individual patients and among different patients with the same type of cancer.
Dr. Jaime Modiano’s lab at the University of Minnesota developed complementary methods to identify more robust, multi-biomarker signatures for cancers using xenografted tumor cells. One method is the use of gene cluster expression summary scores that account for coordinated regulation of multiple genes, overcoming deficiencies of single biomarkers. The second is the use of reconstructed hybrid genomes (mouse host and tumor donor species) and bioinformatics approaches to identify mRNAs expressed exclusively in the tumor cells (donor) and mRNAs expressed exclusively in supporting stromal cells (host). These methods can be used for virtually any tumor for which there are cell lines or patient isolates that grow as xenografts (which reflect disease variability). An added benefit of these methods is that they utilize RNA isolated from exosomes. Exosomes can be powerful diagnostic tools. Specifically, they are stable in biological fluids (facilitating easy sample collection through a simple blood draw), can be efficiently isolated, and contain cargo that can be significantly associated with disease states.
Phase of Development
TRL: 3This approach has been validated through identification of a biomarker for residual osteosarcoma in dogs.
Desired Partnerships
This technology is now available for:- License
- Sponsored research
- Co-development
Please contact our office to share your business’ needs and learn more.
Researchers
- Jaime Modiano, VMD, PhD Perlman Professor of Oncology and Comparative Medicine, Department of Veterinary Clinical Sciences
-
expand_more library_books References (1)
- Milcah C. Scott, et al. (2016), Heterotypic mouse models of canine osteosarcoma recapitulate tumor heterogeneity and biological behavior, Disease Models & Mechanisms
-
expand_more cloud_download Supporting documents (1)Product brochureA method to identify cell-free biomarkers of cancers and other diseases.pdf