Plasma cell-free DNA sequencing assay for advanced prostate cancer
A liquid biopsy assay for predicting survival outcomes of castration-resistant prostate cancer
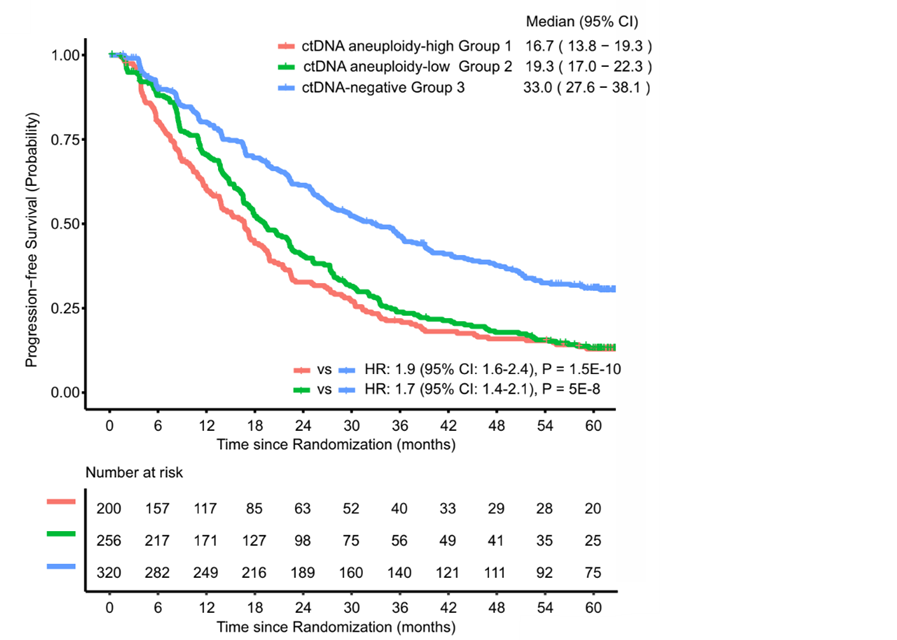
Applications
- Prognosis of castration-resistant prostate cancer
- Monitoring phase III clinical trials with androgen receptor inhibitors
Key Benefits & Differentiators
- Increased sensitivity: Detects critical molecular alterations that occur even in otherwise ctDNA-negative patients
- Informs Treatment: Enables identification of key molecular alterations in targeted genes
Technology Overview
Standard of care treatments for patients with advanced prostate cancer include therapies that inhibit the androgen receptor (AR). However, the disease will eventually progress to castration resistant prostate cancer (CRPC), which is a disease state responsible for nearly all prostate cancer deaths. 20-30% of CRPC patients have primary resistance to AR-targeted therapies, necessitating AR-independent therapies and prognostic biomarkers to identify said resistance for optimizing treatment regimes and designing efficient clinical trials. Previous studies have provided evidence for clinical utility of analysis of cell-free DNA (cfDNA) isolated from plasma of men with metastatic CRPC (mCRPC), with detection of circulating tumor DNA (ctDNA) in the cfDNA in particular associated with poor clinical outcomes. Commercial cfDNA assays are available for identifying patients with ctDNA, and detecting a subset of the genomic alterations that underlie CRPC-specific resistance mechanisms. However, there is an unmet need for cfDNA assays able to prognosticate outcomes in mCRPC patients, particularly in the context of AR-targeted therapies.
Researchers at the University of Minnesota have developed a liquid biopsy assay for predicting survival outcomes of mCRPC patients being treated with AR inhibitors. Specifically, this assay is a DNA sequencing assay for detecting ctDNA in plasma cell-free DNA with specific targeting of mCRPC-specific alterations. Out of 776 cfDNA samples analyzed, 324 cfDNA samples (~42%) were identified as ctDNA-positive using standard parameters measured by commercial DNA sequencing assays, while this new assay identified an additional 156 ctDNA-positive samples. This represents a 20% increase in sensitivity by accounting for mCRPC specific alterations. This additional information can also be leveraged to adapt treatment plans.
Phase of Development
TRL: 3-4Research-grade prototype of assay developed and tested in plasma samples from 776 patients.
Desired Partnerships
This technology is now available for:- License
- Sponsored research
- Co-development
Please contact our office to share your business’ needs and learn more.
Press Releases
University of Minnesota Medical School 12/11/2024Researchers
- Scott Dehm, PhD Professor, Department of Laboratory Medicine & Pathology
- Todd Knutson, PhD Research Manager, Minnesota Supercomputing Institute
-
expand_more library_books References (2)
- Todd P. Knutson, Bin Luo, Anna Kobilka, Jacqueline Lyman, Siyuan Guo, Sarah A. Munro, Yingming Li, Rakesh Heer, Luke Gaughan, Michael J. Morris, Himisha Beltran, Charles J. Ryan, Emmanuel S. Antonarakis, Andrew J. Armstrong, Susan Halabi, Scott M. Dehm (2024), AR alterations inform circulating tumor DNA detection in metastatic castration resistant prostate cancer patients, Nature Communications, 15, 10648
- Halabi S, Guo S, Luo B, Yu C, Knutson TP, Kobilka A, Lyman J, Beltran H, Antonarakis ES, Galsky MD, Rosenberg JE, Ryan CJ, Small EJ, Kelly WK, Morris MJ, Page D, Dehm SM, Armstrong AJ (2025 Aug 6), Pathogenic Genomic Alterations in Circulating Tumor DNA Predict Overall Survival in Men with Metastatic Castrate-resistant Prostate Cancer, European Urology
-
expand_more cloud_download Supporting documents (1)Product brochurePlasma cell-free DNA sequencing assay for advanced prostate cancer.pdf