MoonTag and plant-derived activation domains for improved genetic engineering
A novel programmable transcriptional activator that can work with plant-derived activation domains to enable precise and robust gene activation in plants.
Applications
- Crop engineering
- Engineered genetic incompatibility
- Biotherapeutics
Key Benefits & Differentiators
- Increased stability: The MoonTag system is more stable in transgenic plants compared to previous systems like SunTag
- Plant-derived activation domains: New activation domains (DREB2, DOF1) outperform VP64 with up to 3 fold higher transcription activation
- Temperature resilience: Functional across a broad range of growth temperatures which is essential for potential field applications.
Technology Overview
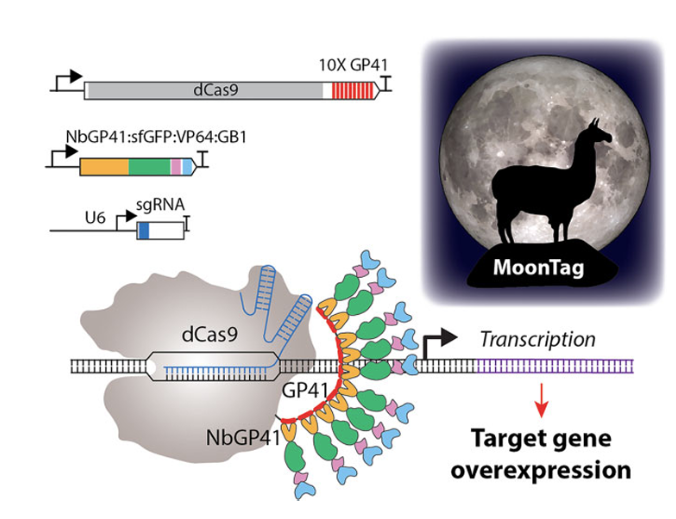
CRISPR-based transcriptional activators have revolutionized gene activation by enabling targeted overexpression of plant genes. SunTag, a second-generation system, activates transcription by recruiting multiple copies of an activation domain to its target promoters. Although SunTag is a strong activator, it is difficult to stably express in some species, thus limiting its applications. Additionally, CRISPR-Cas activators have traditionally relied on non-plant-derived activation domains, such as VP64, which has limited activation efficiency and poses potential regulatory challenges.
To address these problems, researchers at the University of Minnesota have developed a robust programmable transcriptional activator, MoonTag, and novel plant-derived activation domains DREB2 and DOF1. MoonTag uses a nanobody NbGP41 and a GP41 peptide pair to mediate the recruitment of the activation domain. MoonTag demonstrates strong performance in monocots (Setaria) and dicots (Arabidopsis and tomato) and functions across a broad temperature range, making it suitable for field applications. By incorporating plant-specific activation domains such as DREB2 and DOF1, MoonTag delivers up to threefold higher gene activation than VP64. With MoonTag’s enhanced stability and the exceptional activity of novel plant-derived activation domains, these tools represent a significant advancement in precise and robust genetic engineering of plants.
Phase of Development
TRL: 3-4Successfully validated in transgenic Arabidopsis plants with observable phenotypic changes. Other species that MoonTag has been demonstrated in include: Setaria (grasses), corn, tomato, tobacco, pennycress, and camelina.
Desired Partnerships
This technology is now available for:- License
- Sponsored research
- Co-development
Please contact our office to share your business’ needs and learn more.
Researchers
- Michael Smanski, PhD Associate Professor, Department of Biochemistry, Molecular Biology, and Biophysics
- Daniel Voytas, PhD McKnight Presidential Endowed Professor, Department of Genetics, Cell Biology, and Development
-
expand_more library_books References (3)
- Matthew H. Zinselmeier, J. Armando Casas-Mollano, Jonathan Cors, Savio S. Ferreira, Daniel F. Voytas, Michael J. Smanski (2025), Towards engineering hybrid incompatibility in plants, Plant Biotechnology Journal
- Matthew H. Zinselmeier, Juan Armando Casas-Mollano, Jonathan Cors, Adam Sychla, Stephen C. Heinsch, Daniel F. Voytas, Michael J. Smanski (2024), Optimized dCas9 programmable transcriptional activators for plants, Plant Biotechnology Journal, 22, 3202-3204
- J Armando Casas-Mollano , Matthew H Zinselmeier , Adam Sychla , Michael J Smanski (2023), Efficient gene activation in plants by the MoonTag programmable transcriptional activator, Nucleic Acids Research, 51, 7083–7093
-
expand_more cloud_download Supporting documents (1)Product brochureMoonTag and plant-derived activation domains for improved genetic engineering.pdf